Presentation
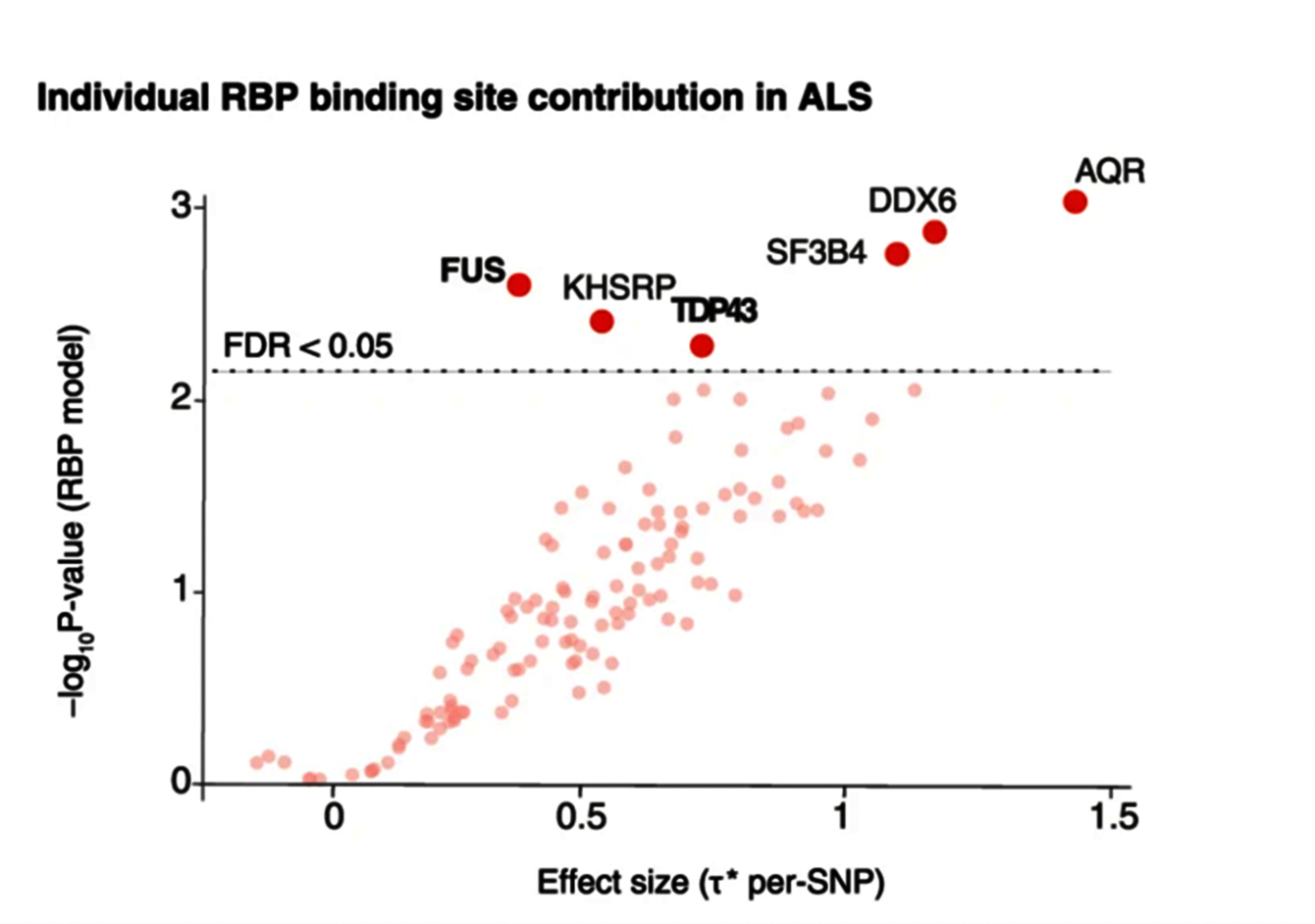
Dot plot showing ALS heritability significantly enriched in subtypes of RNA-binding protein binding sites such as FUS and TDP43 (Megat et al., 2023)
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that manifests as a progressive muscular weakness and paralysis, leading to death within only two to three years upon symptom onset. ALS results from the progressive degeneration of two neuronal populations involved in voluntary motor control: the glutamatergic corticospinal neurons (CSN, or upper motor neurons) in the motor cortex, and the cholinergic motoneurons (MN, or lower motoneurons), in the brainstem and spinal cord. Whereas the vast majority of patients are considered sporadic, about 10% have a familial history, and causative genes have been identified in more than half of these cases, the most represented being C9orf72-SMCR8 complex subunit (C9orf72), Superoxide Dismutase 1 (SOD1), TAR DNA Binding Protein (TARDBP) and FUS RNA binding protein (FUS). While ALS genetics is complex, causative mutations remarkably converge on recurrent dysregulated pathways including DNA repair, RNA and protein metabolism, intracellular trafficking and mitochondrial dysfunction.
In our team we use multi-omics data integration using single nuclei RNAseq from vulnerable and resilient neurons as well as large-scale genetic datasets to discover new risk and protective factors associated with ALS/FTD. Using iPSC-derived motoneurons we aim at validating our new target genes and gain a better understanding on the mechanisms underlying neuroprotection or neurodegeneration in ALS.







